A common question that may cross your mind when you first hear the word ‘epigenetics’ is: where does the word come from? The history of epigenetics is linked to the study of evolution and development [1]. The term was originally coined in 1942 by – the embryologist – Dr. Waddington, and stems from the greek word ‘epigenesis’, which originally described genetic processes’ impact on development [2]. Although the term was not around until the 1940s, the ‘epi’ in ‘epigenetics’, is a concept that can be traced back to the nineteenth century, the notion that genes are influenced by factors beyond the genome [3].
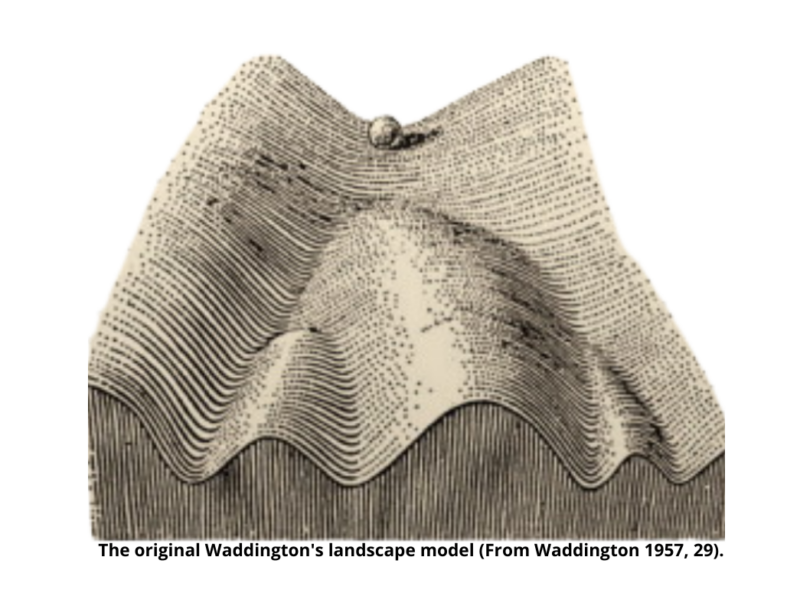
Up until the 1950s, the word ‘epigenetics’ was inaccurately used to include all the developmental events, starting with the fertilized zygote to the mature organism [1]. Around that time, Waddington had proposed the famous epigenetic landscape. He described the cell as a ball that could follow different paths due to the roughness of the surface, symbolizing intra- and extracellular environmental influences [4].
In the period of 1970s-1980s, the identification of high mobility group (HMG) proteins allowed scientists to realize that besides histones (discovered in early 1900s), there are other proteins that play a role in phenotype expression, and in chromatin architecture [5]. Additionally, that was followed by the discovery of the imprinted genes phenomenon, and their regulation according to maternal and paternal inheritance [6].
Nevertheless, DNA methylation (5mC) and post-translational histone modifications were identified shortly after the DNA double helical structure was resolved. DNA methylation was identified early on, in 1965. Other modifications including histone methylation, acetylation, phosphorylation, ubiquitylation, sumoylation and ADP ribosylation were discovered between 1962 and 1977 [8,9].
Despite the fact that quite a bit was understood on DNA and its general organization half way through the last century, epigenetics became extensively studied only in the 1990s and 2000s. Advancements in the field of cloning (e.g. Dolly the sheep) helped to address several unanswered questions [7]. The development of toolkits and biochemical techniques enabled researchers to identify specific enzymes, readers, writers and erasers of epigenetic marks.
As of now, DNA methylation is one of the most studied and well-characterized epigenetic modifications. An example of the importance of this process and its biological relevance is X-inactivation. It is a process by which one of the two X chromosomes is inactivated in therian female mammals. The inactive X chromosome is silenced and packaged into heterochromatin, a structure that is transcriptionally inactive [10]. Hence, silencing takes place due to high levels of DNA methylation accompanied by other histone modifications [11].
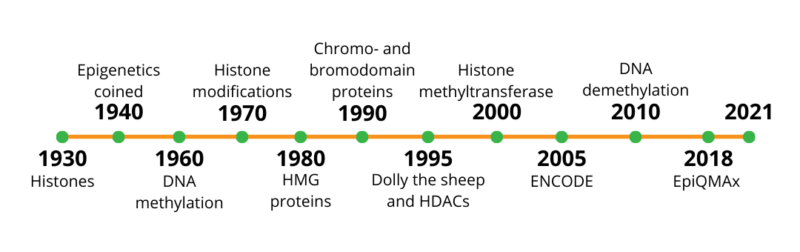
The journey towards a full understanding of the significance of epigenetics is long. Yet, there is a lot to be discovered in this relatively new and exciting field of study. At EpiQMAx, our goal is to help deepen the knowledge on our epigenetic code, validate biomarkers of various diseases and identify new epigenetic drugs. In the near future, EpiQMAx aims to leave a remarkable footprint in this field.

