Have you ever wondered why some of our bodies react differently than others even when performing the exact same exercise? It has been shown that genetic make-up plays a major role when having untrained individuals perform certain physical activities, some will do better while others will do worse [1].
Transcriptomics (study of RNA transcripts) and proteomics (study of proteins) studies have shown that our gene expression patterns adapt to the type of exercise we are doing [1]. For example, individuals who train their endurance have appeared to have different gene expression patterns in skeletal muscles than individuals who train their strength [1]. But not all of it is about our genetic code.
To get a closer look on how exercising results in molecular and systemic effects, an important aspect to account for is epigenetics. Physical exercise was shown to have an impact on histone modifications in muscle and brain tissue, as well as on DNA methylation status in muscle tissue [2, 3]. For example, it has been suggested that physical activity results in DNA methylation of genes that are involved in chronic inflammation and tumor suppression [4].
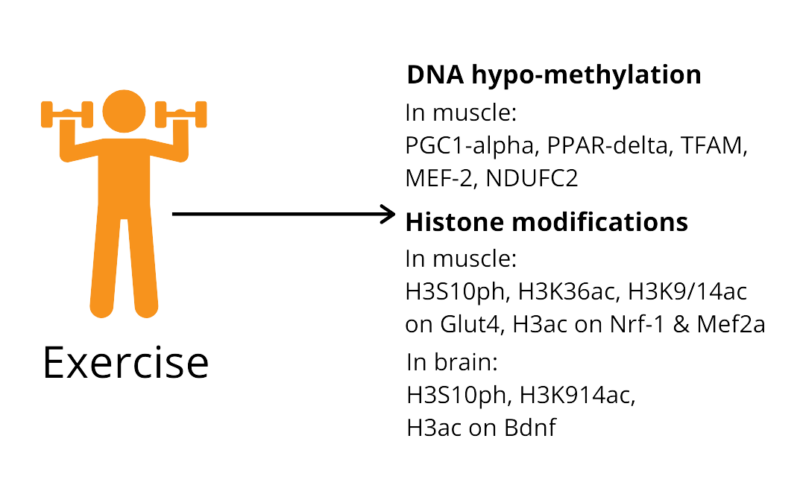
Back to the skeletal muscles example, histone deacetylases (HDAC), enzymes that remove an acetyl group from a histone, are primarily responsible for the epigenetic modifications of histones there [2]. HDACs work on repressing the myocyte enhancer factor 2 (MEF-2) transcription factor, which in return suppresses the formation of slow twitch oxidative myofibers [2]. However, overexpressing MEF-2, through selective degradation of HDACs, in mice skeletal muscle promoted the formation of slow myofibers and enhanced the running endurance, enabling those transgenic mice to run almost twice the distance of the wild type mice [2].
Other studies have shown that exercise increases glucose transporter (GLUT4) gene and protein expression in human skeletal muscle, which generally enhances whole-body insulin action [5]. After exercise, nuclear HDACs were decreased by more than half, and a decrease of more than a quarter of HDACs association with MEF-2 was observed [5].
There are several epigenetic modifications that have been correlated with physical activity. Hence, highlighting potential therapeutic targets for several disorders (e.g. regulation of GLUT4 expression for disorders like diabetes type 2). Together with partners, EpiQMAx aims to deepen our understanding of the effect that physical activity has on us and our epigenetic traits.

